
Therefore, set the number of outputs of the minibatchqueue object to 4. The preprocessMiniBatch function returns 4 variables as output. Use the custom mini-batch preprocessing function preprocessMiniBatch, defined in the Preprocess Mini-Batch Function section of the example to remove zero paddings from the data, compute the number of nodes per graph, and merge multiple graph instances into a single graph instance. If your hardware does not have enough memory, then reduce the mini-batch size.
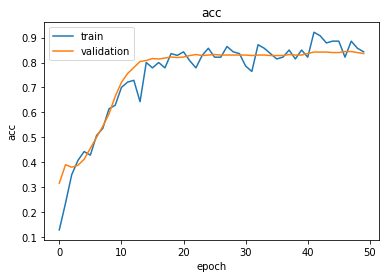
Large mini-batches of training data for GATs can cause out-of-memory errors. Train for 70 epochs with a mini-batch size of 300. Define Model Loss FunctionĬreate the function modelLoss, defined in the Model Loss Function section of the example, which takes as input the model parameters, a mini-batch of input features and corresponding adjacency matrix, the number of nodes per graph, and the corresponding encoded targets of the labels, and returns the loss, the gradients of the loss with respect to the learnable parameters, and the model predictions. This figure illustrates the multilabel graph classification workflow in this example.Ĭreate the function model, defined in the Model Function section of the example, which takes as input the model parameters, the input features and adjacency matrix, and the number of nodes per graph, and returns predictions for the labels. This example considers the functional groups CH, CH2, CH3, N, NH, NH2, NOH, and OH. Each functional group represents a subgraph, so a graph can have more than one label or no label if the molecule representing the graph does not have a functional group. Graph labels are functional groups or specific groups of atoms that play important roles in the formation of molecules. Three physicochemical properties of the atoms are used as node information: scalar Hirshfeld dipole moments, atomic polarizabilities, and Van der Waals radii. The data set contains 5 unique atoms: carbon (C), hydrogen (H), nitrogen (N), oxygen (O), and sulfur (S). Each molecule is composed of up to 23 atoms, which are represented as nodes.
#Graph attention networks how to#
This example shows how to train a GAT using the QM7-X data set, a collection of graphs that represent 6950 molecules. The output features are used to classify the graph usually after employing a readout, or a graph pooling, operation to aggregate or summarize the output features of the nodes. Using the graph structure and available information on graph nodes, GAT uses a masked multihead self-attention mechanism to aggregate features across neighboring nodes, and computes output features or embeddings for each node in the graph. If the observations in your data have a graph structure with multiple independent labels, you can use a GAT to predict labels for observations with unknown labels.
This example shows how to classify graphs that have multiple independent labels using graph attention networks (GATs).


 0 kommentar(er)
0 kommentar(er)
